vaex: lazy, parallel dataframes
When inspecting and manipulating data in Python, Pandas has been the defacto solution as it’s intuitive and works for most cases. One of the cases which it starts to falter on is “large” datasets. To address this, there are many solutions. Some, like dask and Spark, schedule work to clusters to distrbute work. Others, like vaex, just load less.
vaex’s solution of loading less can be done similarly with Pandas but it does so while presenting the facade of loading the entire dataset and without the need for additional nodes. This is great because distributed systems are hard and fragile.
Loading Data
Working with a generated random dataframe with a timestamp column and 3 double-precision float columns, loading 5.2M rows in various file formats:
# ~1.63GB file on disk
timeit df = vaex.open('/home/azureuser/dev/test.parquet')
# 1.11 ms ± 31.8 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
df.schema()
# {'ts': datetime64[ns], 'bid': float64, 'ask': float64, 'close': float64}
# ~1.68GB file on disk (arrow IPC format)
timeit df = vaex.open('/home/azureuser/dev/test.arrow')
# 6.39 ms ± 323 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
# ~1.49GB file on disk (v2 format, lz4 compressed)
timeit df = vaex.open('/home/azureuser/dev/test.feather')
# The slowest run took 16.85 times longer than the fastest. This could mean that an intermediate result is being cached.
# 4.03 s ± 5.01 s per loop (mean ± std. dev. of 7 runs, 1 loop each)
# ~1.68GB file on disk (v2 format, uncompressed)
timeit df = vaex.open('/home/azureuser/dev/test-uncompressed.feather')
# 27.7 ms ± 741 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
# ~4.09GB file on disk
timeit df = vaex.open('/home/azureuser/dev/test.csv')
# crashed 2CPU/8GB VM
timeit df = pandas.read_parquet('/home/azureuser/dev/test.parquet')
# 1.84 s ± 66 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Loading the same Parquet file in vaex is done in a fraction of the time compared to the equivalent command in Pandas. It should be noted, that loading a compressed feather file in vaex appears to load the entire dataset into memory as well.
The memory usage of importing the vaex library adds ~75MB but when working with Arrow or Parquet files, loading a ~1.6GB file results in a ~60MB increase in memory.
Grouping Data
A common use case given a time-series dataset is to group and aggregate the data. In a prior post done years ago, I found Gnocchi did a decent job at this. For those unfamiliar with Gnocchi, it only builds series and not dataframes so it provides less functionality and flexibility but this is only highlighting vaex’s grouping ability.
Using Gnocchi’s time-series functionality:
from gnocchi import carbonara
import numpy as np
dates = np.arange("2000-01-01", "2100-01-01", dtype='datetime64[m]')
g_ts = carbonara.make_timeseries(dates, numpy.random.rand(len(dates)))
#array([('2000-01-01T00:00:00.000000000', 0.99583813),
# ('2000-01-01T00:01:00.000000000', 0.43503748),
# ('2000-01-01T00:02:00.000000000', 0.39603606), ...,
# ('2099-12-31T23:57:00.000000000', 0.19333233),
# ('2099-12-31T23:58:00.000000000', 0.80041872),
# ('2099-12-31T23:59:00.000000000', 0.5249474 )],
# dtype=[('timestamps', '<M8[ns]'), ('values', '<f8')])
timeit carbonara.GroupedTimeSeries(g_ts, 'M').mean()
# 4.05 s ± 9.53 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
timeit carbonara.GroupedTimeSeries(g_ts, 'M').max()
# 18.6 s ± 79.6 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
timeit carbonara.GroupedTimeSeries(g_ts, 'M').count()
# 3.41 s ± 11.1 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
It’s able to group 5.2M datapoints to 1.2K groups between ~3.5s and ~20s depending on the aggregation method. Doing the same with vaex:
import vaex
df = vaex.open('/home/azureuser/dev/test.parquet')
## ts bid ask close
#0 datetime.datetime(2000, 1, 1, 0, 0) 0.464756415970771 0.5034086333555521 0.08377915113368695
#1 datetime.datetime(2000, 1, 1, 0, 1) 0.829357151822713 0.06565148686200395 0.6484271256241894
#2 datetime.datetime(2000, 1, 1, 0, 2) 0.5923924050137601 0.4438634343436415 0.5739442873602649
#3 datetime.datetime(2000, 1, 1, 0, 3) 0.03331627430636752 0.541054600412977 0.9770609492344751
#4 datetime.datetime(2000, 1, 1, 0, 4) 0.13478689648054032 0.9920226087831646 0.28975132591720887
#... ... ... ... ...
#52,595,995 datetime.datetime(2099, 12, 31, 23, 55) 0.5916867847527888 0.5695370700198127 0.42545814565311113
#52,595,996 datetime.datetime(2099, 12, 31, 23, 56) 0.527014400353311 0.16559317797248707 0.9624458703937147
#52,595,997 datetime.datetime(2099, 12, 31, 23, 57) 0.7288556992043 0.5565629760578215 0.3842203259165041
#52,595,998 datetime.datetime(2099, 12, 31, 23, 58) 0.6451698531740229 0.9911780871685453 0.3645913500554936
#52,595,999 datetime.datetime(2099, 12, 31, 23, 59) 0.5830517027666722 0.7555986017849898 0.49501895044487865
timeit group_per_month = df.groupby(
by=vaex.BinnerTime(df.ts.astype('datetime64[ms]'), resolution='M'),
agg={'mean': vaex.agg.mean(df.bid)})
# 3.34 s ± 40.4 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
timeit group_per_month = df.groupby(
by=vaex.BinnerTime(df.ts.astype('datetime64[ms]'), resolution='M'),
agg={'max': vaex.agg.max(df.bid)})
# 2.95 s ± 33.2 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
timeit group_per_month = df.groupby(
by=vaex.BinnerTime(df.ts.astype('datetime64[ms]'), resolution='M'),
agg={'count': vaex.agg.count(df.bid)})
# 2.98 s ± 31.6 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
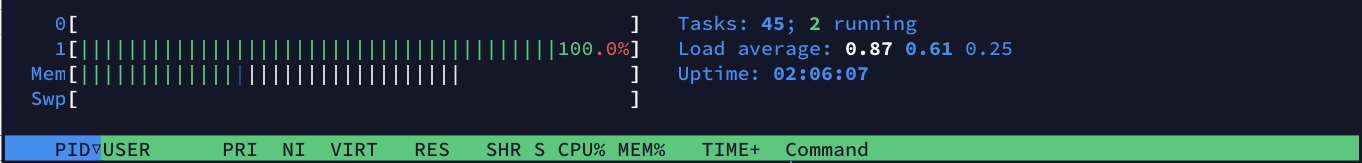 gnocchi
gnocchi
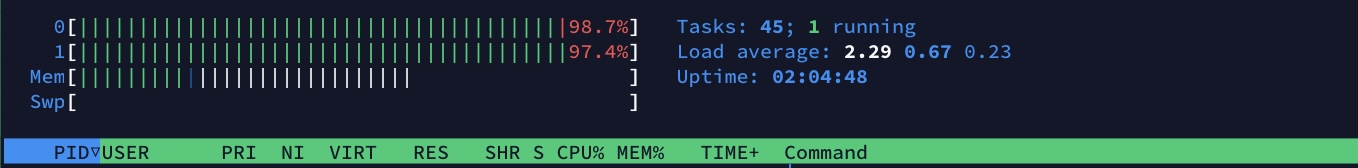 vaex
vaex
It is able to aggregate faster in all cases by parallelising the operation and making use of all available cores where as Gnocchi treats the entire series using a single thread. vaex does this while providing a more functional interface.
Additionally, vaex required ~0.8GB of memory to perform the aggregation (solely attributed to BinnerTime) while doing the same aggregation in Gnocchi required up to 1.3GB of memory in addition to that required to load the entire initial dataset into memory.
When dealing with large datasets, vaex presents a viable solution for data inspection.
ps. or maybe polars is better?